Constantine publishes nucleation-based self-assembly for pattern recognition in Nature
Group member Constantine Evans, and his collaborators Jackson O’Brien, Erik Winfree and Arvind Murugan, published a beautiful self-assembly paper in Nature this week that uses patterns of high and low concentration tiles to kinetically control self-assembly of one of several shapes.
Here's my quick take, best savoured with a nice espresso, as is Constantine's tipple. By the way, I highly recommend Andrew Phillips' very nice News and Views article for an overview, or some local hot takes.
The gist of it goes like this. Their tile system has three, overlapping, tilesets that share some tiles: if all tiles are at equal concentration a simple temperature anneal yields a mixture of three shapes at similar concentrations (part a of the image below).
But they show one can bias the mixture to assemble one target shape out of three by simply having tiles at high concentration in a small region of the target shape called the flag, and all other tiles at low concentration. The clever part is that although flag tiles appear in the other two shapes also, they are quite spread out, so when assembly happens while cooling the system down, there is a large kinetic drive towards the target, and a negative feedback that starves the other two shapes. As a result we see one, and only one, shape.
To my mind, this has the potential to be an incredibly clean and neat way to seed DNA tile assemblies, without the bespoke DNA origami seed nanostructures used in prior work including our own.
But they don't stop there. Next, they go beyond the flag idea to have a more complex notion of concentration pattern, best described as a concentration vector (each tile has a potentially unique concentration). Depending on the vector, we get one of three shapes (typically). But vectors can encode data, just like bitstrings or the letters you're reading. So, using a python program, Constantine, Jackson, Erik and Arvind map pictures of horses, apples, and scientists to concentration vectors and, via that encoding, their system recognises and characterises images, even if those images are corrupted by noise (see part b below).
We've known for years that self-assembly based growth processes can run algorithms, now we've learned that the process of nucleation can to, albeit rather differently from anything we've seen thus far!
Really cool. Check out the full paper for more details.
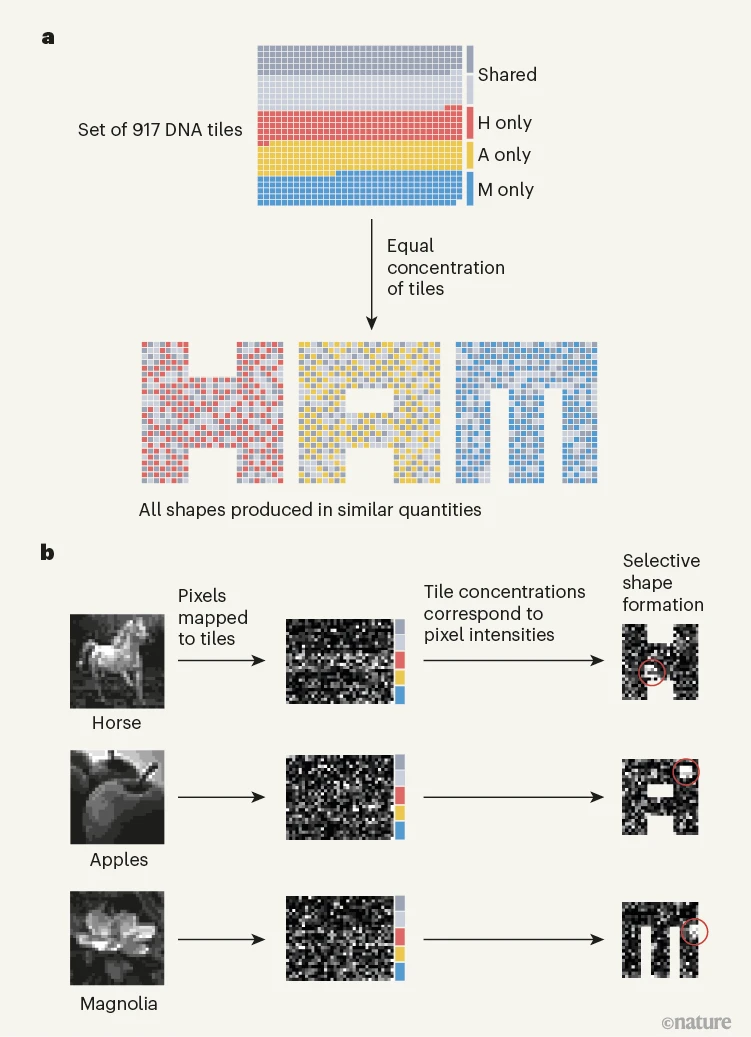
Image taken from Andrew Phillips' News and Views, copyright Nature, which was adapted from Evans et al.