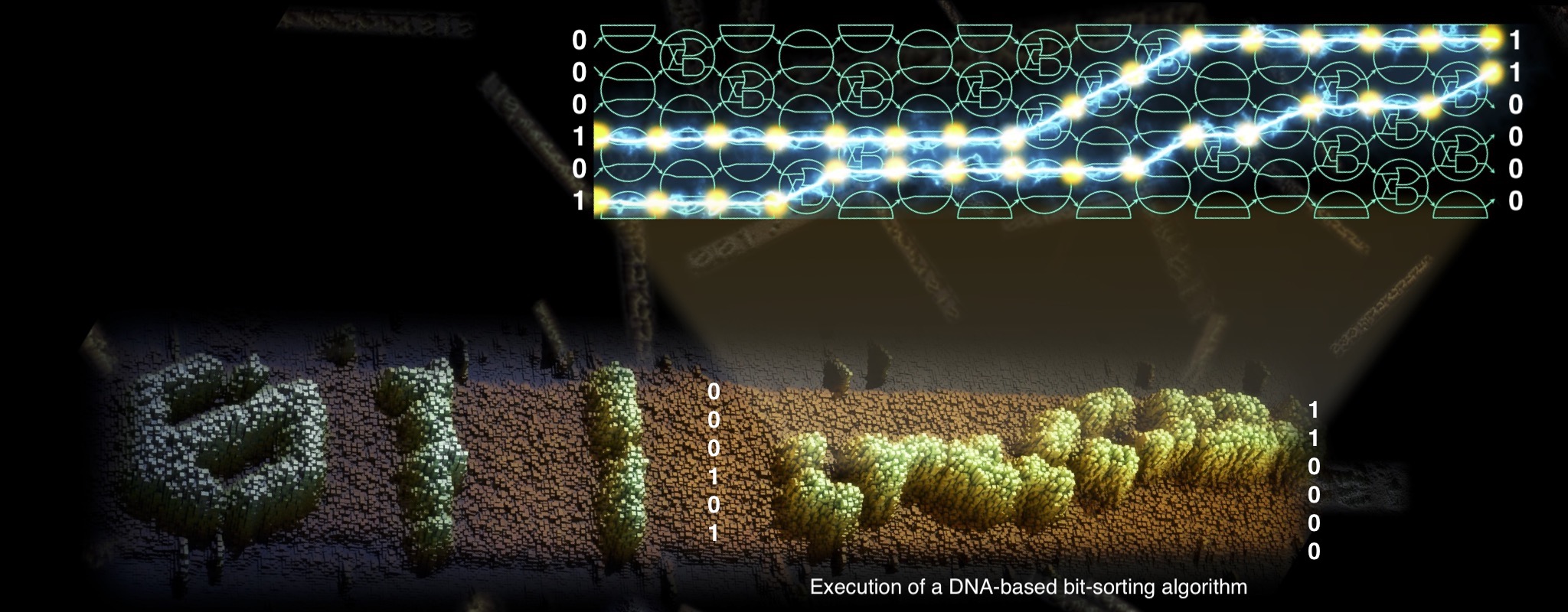
TAPDANCE: Theory And Practice of DNA Computing Engines
We carry out fundamental research on molecular computers: collections of carefully engineered DNA strands that interact and bump into each other in a test tube to solve computational problems. Our focus is on both the underlying computational theory and practical implementation in the lab. Based at the Hamilton Institute, Maynooth (Ireland), and funded by ERC, EIC, and SFI.
Latest news: blog posts
- Visitors to our group in 2025
- Welcoming Josh, Ashley, Eoghan, Oliver, and Daniel
- ICALP best paper award for Ahmed's multistrand MFE algorithm
- A PhD in Reconfigurable Molecular Robots! Congrats to Cai!
- Abeer wins Research Ireland Pathways award
- Constantine awarded the Dirks prize!
- BB(5) makes Scientific American top 7 of 2024!
- Shinnosuke Seki visit
- Positions availiable — Postdoc, Senior Postdoc, Technical Officer, Senior Technical Officer
- DISCO project launch
- Igor Potapov visit
- Positions availiable — Postdoc, Senior Postdoc, Technical Officer, Senior Technical Officer
- The 5th busy Beaver number is solved! BB(5) = 47,176,870. Congrats Tristan et al!
- Welcome new group members!
- Sergiu Ivanov joins us for his sabbatical
- 2x Hamilton Prof jobs
- Sarah Guerin seminar and visit
- Visitor Damien Baigl joins us for a few days
- Constantine publishes nucleation-based self-assembly for pattern recognition in Nature
- 6 tiles are all you need for a PhD! Congrats to Tristan!
- We're off to DNA28!
- Welcome new group members!
- Trent's work funded by SFI
- Turning Machines — a simple algorithmic model for molecular robotics
- Group funding from SFI
- Molpigs interview with Damien
- Can we know busy beaver value BB(15)? — new paper
- New postdoc Abeer Eshra joins our group!
- Computing with small tile sets — new paper at DNA27
- New postdoc Ismael Mullor Ruiz joins our group!
- Waiting for Gödel — Tristan talks about Turing machines, Godel encodings, undecidability and incompleteness
- Ulysse Léchine joins our group!
- Positions availiable — Postdoc, Senior Postdoc and Technical officer
- Video and slides for keynote talk at DNA26
- Turning machines! New paper on reconfigurable robotics
- Cai Wood joins our group!
- The Collatz problem embeds a base conversion algorithm — new paper
- The Collatz problem and regular expressions — new paper by Tristan
- STOC 2020 video presentation by Pierre on pumpablity in self-assembly
- SFI award for medium-term Covid-19 forecasting
- Constantine's video presentation for FNANO 2020
- Covid-19 dashboard
- Paper proving pumpability in noncooperative self-assembly!
- New postdoc Constantine Evans
- Slides on DNA-based algorithmic self-assembly of 6-bit iterated Boolean circuits
- Sabbatical visitor Prof Matthew Patitz
- Codenano, a tool for designing DNA nanostructures
- New postdoc Trent Rogers
- New summer students Eilbhe, Dara, Cai
- New interns Nicolas Levy and Coline Petit-Jean
- Assembling large structures without cooperation, published at DNA25
-
Paper on reprogrammable DNA self-assembly, published in Nature
See the paper and media coverage.
- New Postdoc & PhD positions
-
Funding from SFI
New SFI award to fund our work
-
Funding from ERC
New ERC Consolidator award to fund our work
- Move to Hamilton Institute at Maynooth University
- Self-assembly workshop at UCNC & MCU
- Tristan wins best poster award at DNA23!
-
Molecular cargo sorting robots, accepted to Science
A molecular robot that sorts tiny cargos into separate piles
-
Pierre-Étienne talks at STOC
Pierre talks about tile self-assembly at STOC 2017; no bounded Turing machine simulation and no intrinsic universality at temperature 1.
-
Papers accepted to DNA23
Two papers accepted to DNA23; experiments on algorithmic self-assembly with DNA tiles, and theory of a new thermodynamic model of computation!
-
Recent talks by Damien
Damien talks about DNA experiments on algorithmic self assembly at the Foundations of Nanoscience conference (FNANO), and DNA-based self-replicators at OCAV (an origin of life working group).
-
Recent talks by Pierre-Étienne
Pierre-Étienne talks about non-cooperative self-assembly at NWC, and Pijul at Rust Belt Rust and Rust Fest.
-
Non-cooperative self-assembly is provably weak
TAPDANCE's first publication, accepted to STOC 2017
-
Teaching at ENS Lyon
Damien teaches a 1-week graduate course at ENS Lyon on molecular computation; theory & experiments.